Medical Working Group

Mission
Develop benchmarks and best practices that help accelerate AI development in healthcare through an open, neutral, and scientific approach.
Purpose
Medical AI has tremendous potential to advance healthcare by supporting the evidence-based practice of medicine, personalizing patient treatment, reducing costs, and improving provider and patient experience. To unlock this potential, we believe that robust evaluation of healthcare AI and efficient development of machine learning code are important catalysts.
To address these two aims the Medical working group’s efforts are geared towards creating the fabric necessary for proper benchmarking of medical AI. This includes: (1) developing the MedPerf platform and GaNDLF to (2) establish a shared set of standards for benchmarking of Medical AI, (3) incorporate and disseminate best practices for our efforts, and (4) create a robust medical AI ecosystem by partnering with key and diverse global stakeholders that span patient groups, academic medical centers, commercial companies, regulatory and oversight bodies, and non-profit organizations.
Within the Medical working group we strongly believe that these efforts will give key stakeholders the confidence to trust models and the data/processes they relied on, therefore accelerating ML adoption in the clinical settings, possibly improving patient outcomes, optimizing healthcare costs, and improving provider experiences.
Standards
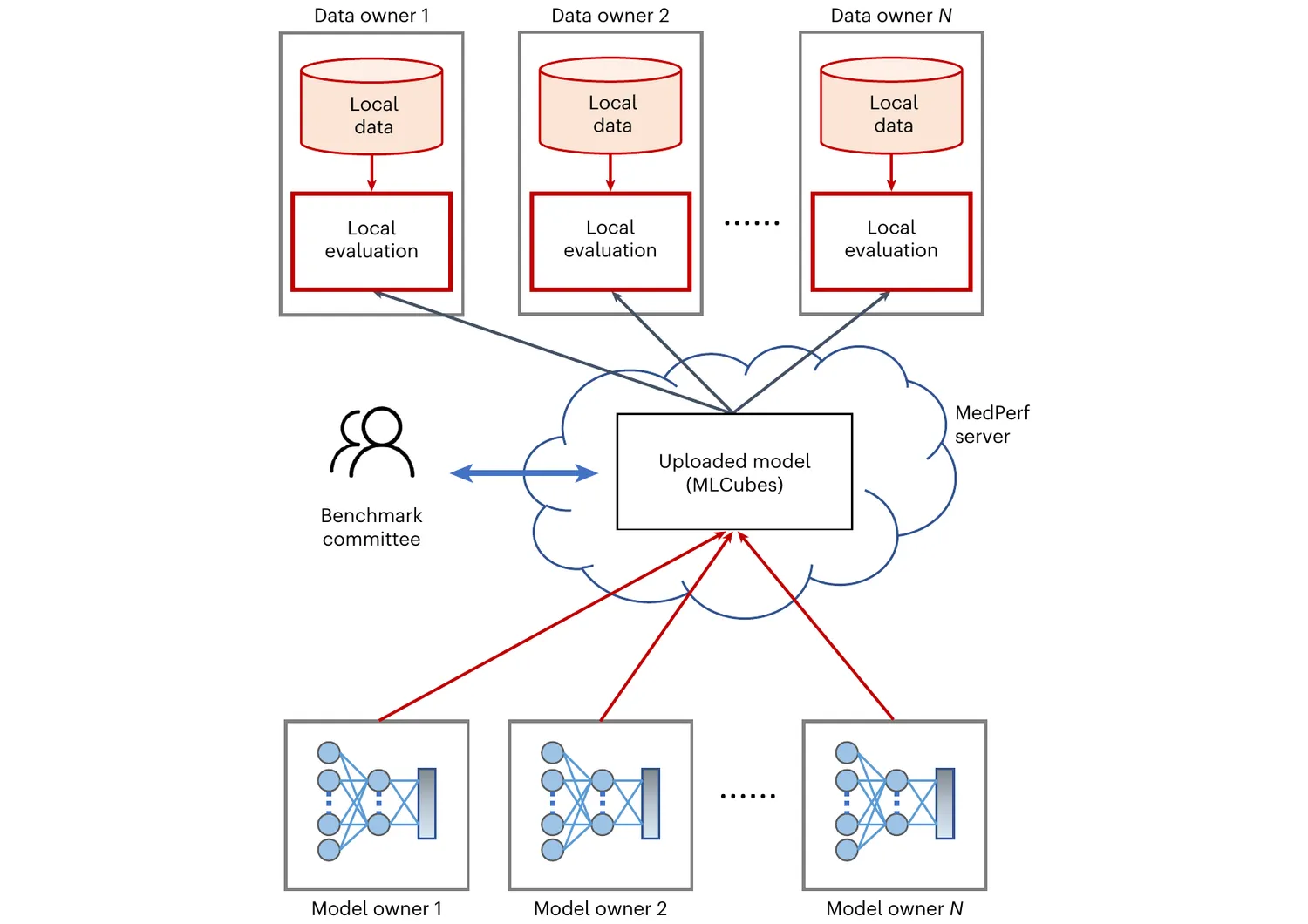
MedPerf is an open benchmarking platform that aims at evaluating AI on real world medical data. MedPerf follows the principle of federated evaluation in which medical data never leaves the premises of data providers. Instead, AI algorithms are deployed within the data providers and evaluated against the benchmark. Results are then manually approved for sharing with the benchmark authority. This effort aims to establish global federated datasets and to develop scientific benchmarks reducing risks of medical AI such as bias, lack of generalizability, and potential misuse. We believe that this two fold strategy will enable clinically impactful AI and drive healthcare efficacy. More information can be found at the MedPerf website and GitHub repository.
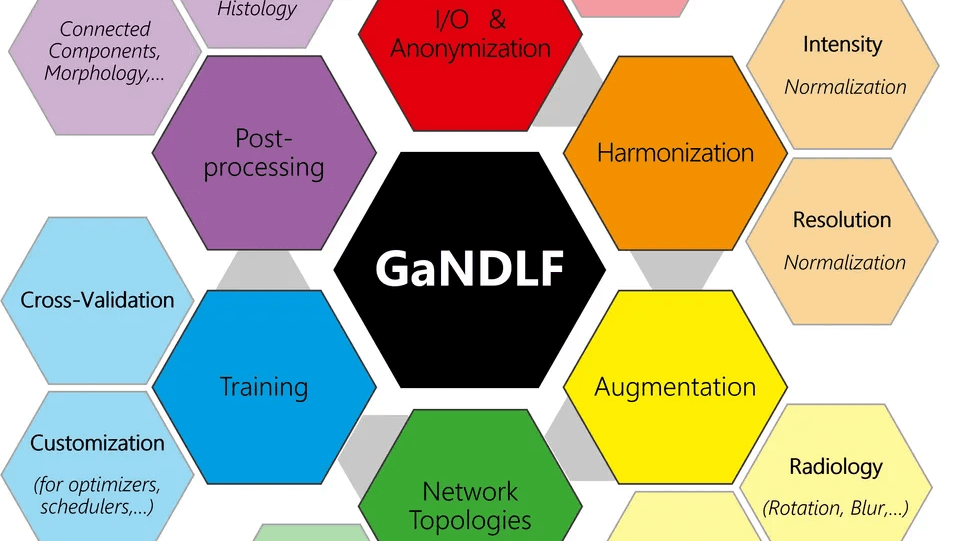
GaNDLF is an open framework for developing machine learning workflows focused on healthcare. It follows zero/low code design principle and allows researchers to train and infer robust AI models without having to write a single line of code. Our effort aims to democratize medical AI through easy to use interfaces, well-validated machine learning ideologies, robust software development strategies, and a modular architecture to ensure integration with multiple open-source efforts. By focusing on end-to-end clinical workflow definition and deployment, we believe GaNDLF will turbocharge the AI landscape in healthcare. More information can be found at the GaNDLF website, GitHub repository, and paper.
Benchmarking of Medical AI
To-date we have worked with professionals from multiple hospitals, companies, universities, and groups:
| A*STAR, Singapore | Microsoft, Redmond, WA, USA |
| Amazon, Seattle, WA, USA | NVIDIA, Santa Clara, CA, USA |
| Brigham and Women’s Hospital, Boston, MA, USA | Nutanix, San Jose, CA, USA |
| Broad Institute of MIT and Harvard, Cambridge, MA, USA | OctoML, Seattle, WA, USA |
| Cisco, San Jose, CA, USA | Perelman School of Medicine, Philadelphia, PA, USA |
| Dana-Farber Cancer Institute, Boston, MA, USA | Red Hat, Raleigh, NC, USA |
| Factored, Palo Alto, CA, USA | Rutgers University, New Brunswick, NJ, USA |
| Flower Labs, Hamburg Germany | Sage Bionetworks, Seattle, WA, USA |
| Fondazione Policlinico Universitario A. Gemelli IRCCS, Rome, Italy | Stanford University School of Medicine, Stanford, CA, USA |
| German Cancer Research Center, Heidelberg, Germany | Stanford University, Stanford, CA, USA |
| Google, Mountain View, CA, USA | Supermicro, San Jose, CA, USA |
| Harvard Medical School, Boston, MA, USA | Tata Medical Center, Kolkata, India |
| Harvard T.H. Chan School of Public Health, Boston, MA, USA | University of Cambridge, Cambridge, UK |
| Harvard University, Cambridge, MA, USA | University of Heidelberg, Heidelberg, Germany |
| Hugging Face, New York, Ny, USA | University of Pennsylvania, Philadelphia, PA, USA |
| IBM Research, San Jose, CA, USA | University of Queensland, Brisbane, Australia |
| IHU Strasbourg, Strasbourg, France | University of Strasbourg, Strasbourg, France |
| Intel, Santa Clara, CA, USA | University of Toronto, Toronto, Canada |
| John Snow Labs, Lewes, DE, USA | University of Trento, Trento, Italy |
| Landing.AI, Palo Alto, CA, USA | University of York, York, UK |
| Lawrence Livermore National Laboratory, Livermore, CA, USA | Vector Institute, Toronto, Canada |
| MLCommons, San Francisco, CA, USA | Weill Cornell Medicine, New York, NY, USA |
| Massachusetts Institute of Technology, Cambridge, MA, USA | Write Choice, Florianópolis, Brazil |
| Meta, Menlo Park, CA, USA | cKnowledge, Paris, France |
| Fast.ai, San Francisco, CA, USA |
Call for Participation
We cannot achieve our goals without the help of the broader technical and medical community. We call for the following:
- Healthcare stakeholders to form benchmark committees that define specifications and oversee analyses.
- Participation of patient advocacy groups in the definition and dissemination of benchmarks.
- AI researchers, to test the end-to-end platform and use it to create and validate their own models across multiple institutions around the globe.
- Data owners (e.g., healthcare organizations, clinicians) to register their data in the platform (while never sharing the data).
- Data model standardization efforts to enable collaboration between institutions, such as the OMOP Common Data Model, possibly leveraging the highly multimodal nature of biomedical data.
- Regulatory bodies to develop medical AI solution approval requirements that include technically robust and standardized guidelines.
Deliverables
- MedPerf
- Improve metadata benchmarking registry
- Interface with common medical data infrastructures (e.g., XNAT)
- Support Federated Learning with 3rd parties
- Research and development of clinically impactful benchmarks (e.g., health equity, bias, dataset diversity)
- Co-develop and support benchmarking efforts
- GaNDLF
- Improved support for non-imaging data types
- Include extensive tutorials for both developers and users
- Add support for cross-device training
- Add more state-of-the-art model architectures that work across different data types
- Improve reported metrics
- Add support for new libraries (PyTorch 2.0, Pandas 2.0) that have API breaks
Meeting Schedule
Last Wednesday of the month from 10:05-11:00AM Pacific
Join
Related Blog
-
Announcing MedPerf Open Benchmarking Platform for Medical AI
Bridging the gap between Medical AI research and real-world clinical impact
-
Launching GaNDLF for Scalable End-to-End Medical AI Workflows
A powerful framework combining AI with medical research to enable medical professionals to more effectively diagnose and treat patients
Medical Working Group Projects
GaNDLF
MedPerf
How to Join and Access Medical Working Group Resources
- To sign up for the group mailing list, receive the meeting invite, and access shared documents and meeting minutes:
- Fill out our subscription form and indicate that you’d like to join the Medical Working Group.
- Associate a Google account with your organizational email address.
- Once your request to join the Medical Working Group is approved, you’ll be able to access the Medical folder in the Public Google Drive.
- To engage in working group discussions, join the group’s channels on the MLCommons Discord server.
- To access the GitHub repositories (public):
- If you want to contribute code, please submit your GitHub ID to our subscription form.
- Visit the GitHub repositories:
Medical Working Group Chairs
To contact all Medical AI working group chairs email [email protected].
Alexandros Karargyris
Alexandros Karargyris has been leading projects related to applications in the intersection of surgery and artificial intelligence (AI). Previously, he worked as a researcher at IBM and NIH for more than 10 years. His research interests lie in the space of medical imaging, machine learning and mobile health. He has contributed to healthcare commercial products and imaging solutions deployed in under-resourced areas. His work has been published in peer-reviewed journals and conferences.
Renato Umeton
Renato Umeton heads artificial intelligence (AI) and data science in the Informatics & Analytics department of Dana-Farber Cancer Institute, a teaching-affiliate of Harvard Medical School. His work focuses on Operations, translating AI from the research realm into the Clinic and into Enterprise software offerings that benefit patients and cancer researchers, at scale. Renato started working on artificial intelligence, data science and big data in 2007, when these areas were yet to be well defined; since then he has published in several scientific fields, he has worked in academia, in consulting, in hospital settings, in biotech, and is currently affiliated also with Harvard School of Public Health, Massachusetts Institute of Technology, and Weill Cornell Medicine.
Vice Chairs
Micah Sheller
Micah Sheller currently works as a senior research scientist in Intel’s Security and Privacy Research Labs, where he leads secure federated learning research. He developed the first version of the OpenFL open-source federated learning platform and is the technical federated learning lead for the FeTS initiative, which recently trained a 3DResUNet across 53 hospitals. Micah has had the pleasure of working on a wide range of projects since his first Intel internship in 1999, when he worked on the Intel Web Tablet. Work in USB 3.0, the prototype Intel SGX software runtime, passive and continuous biometrics and more has kept Micah happily learning and making friends throughout his career.
Spyridon Bakas
Vice Chair for Benchmarking & Clinical Translation
Spyridon Bakas is an Assistant Professor at the Perelman School of Medicine at the University of Pennsylvania, focusing on the development, application, and benchmarking of computational algorithms in medical imaging, with the intention of improving disease assessment and diagnosis in the current clinical practice. He has been leading projects on image quantification, radiogenomics, and federated learning, towards enabling treatment selection models customized on an individual patient basis, while addressing health disparities and inequities. He has published in numerous peer-reviewed journals and conferences, he is a board member of the MICCAI Society’s Special Interest Group on Biomedical Image Analysis Challenges (SIG-BIAS) and has served as the organizer and chair of numerous computational challenges, workshops, and tutorials at both technical and clinical scientific meetings.
Technical Leads
Johnu George
Johnu George is a staff engineer at Nutanix with a wealth of experience in building production grade cloud native platforms. He has a strong distributed systems background and has led efforts in building large scale hybrid data pipelines. He holds multiple patents in this area and has been an invited speaker at various conferences like Kubecon, Apache Big Data etc. He is an active open source contributor and has steered several industry collaborations on projects like Kubeflow, Apache Mnemonic and Knative. His current research interests include machine learning system design, distributed learning infrastructure improvements and ML workloads characterization. He is an Apache PMC member and currently chairing Kubeflow Training and AutoML Working groups.
Alejandro Aristizabal
Alejandro Aristizabal is a machine learing engineer with Factored. He has more than 4 years of experience in Software Development. He holds a bachelors degree in sound engineering. He is very interested in AI Safety, Security and Privacy, as well as human-machine interaction.
Sarthak Pati
Sarthak Pati
Sarthak Pati is a researcher and software engineer with a focus on artificial intelligence and clinical workflow management. He holds a Bachelor’s degree in Biomedical Engineering from Manipal University (India), and a Master’s degree in Biomedical Computing from the Technical University of Munich (Germany). During his time at UPenn, he has been working in image analysis, machine learning, and federated learning for medicine, and has led and contributed to the development of large open-source projects, such as the Federated Tumor Segmentation (FeTS) Tool, the Generally Nuanced Deep Learning Framework (GaNDLF), and the Open Federated Learning (OpenFL) library. In his free time, he enjoys hiking with his dog, Mocha.
Hasan Kassem
Hasan Kassem is a machine learning engineer with a background in MLOps and Federated Learning. He has been working for more than three years in the fields of software engineering and AI. He holds a master’s degree in robotics and intelligent systems, where his focus was on federated machine learning for surgical applications. He enjoys his free time playing chess and understanding the psychology of human personalities.
Theme Leads
Prakash Narayana Moorthy
Prakash Narayana Moorthy is a research software engineer at the Security and Privacy research labs, Intel Corporation, where he has been working on the design and implementation of trustworthy distrusted system for the past 3 years. Prakash currently works on architectures for privacy-preserving smart contract platforms, exploring the role of blockchains in establishing trustworthy federated learning pipelines, and also acts as a security architect for the OpenFL federated learning platform. Prior to joining Intel, Prakash was a postdoc at the EECS department, Massachusetts Institute of Technology, where he invented multiple algorithms for consistent distributed data storage. Prakash holds a Ph.D. in electrical engineering from Indian Institute of Science, Bangalore. Past industry experiences include multiple internships with NetApp as well as working for more than 2 years as a wireless communications engineer for Beceem, a 4G fabless semiconductor company that got acquired by Broadcom.