Medical AI
Develop benchmarks and best practices that help accelerate AI development in healthcare through an open, neutral, and scientific approach.

Purpose
Medical AI has tremendous potential to advance healthcare by supporting the evidence-based practice of medicine, personalizing patient treatment, reducing costs, and improving provider and patient experience. To unlock this potential, we believe that robust evaluation of healthcare AI and efficient development of machine learning code are important catalysts.
To address these two aims the Medical AI working group’s efforts are geared towards creating the fabric necessary for proper benchmarking of medical AI. This includes: (1) developing the MedPerf platform and GaNDLF to (2) establish a shared set of standards for benchmarking of Medical AI, (3) incorporate and disseminate best practices for our efforts, and (4) create a robust medical AI ecosystem by partnering with key and diverse global stakeholders that span patient groups, academic medical centers, commercial companies, regulatory and oversight bodies, and non-profit organizations.
Within the Medical AI working group we strongly believe that these efforts will give key stakeholders the confidence to trust models and the data/processes they relied on, therefore accelerating ML adoption in the clinical settings, possibly improving patient outcomes, optimizing healthcare costs, and improving provider experiences.
Deliverables
MedPerf
- Improve metadata benchmarking registry
- Interface with common medical data infrastructures (e.g., XNAT)
- Support Federated Learning with 3rd parties
- Research and development of clinically impactful benchmarks (e.g., health equity, bias, dataset diversity)
- Co-develop and support benchmarking efforts
GaNDLF
- Improve metadata benchmarking registry
- Interface with common medical data infrastructures (e.g., XNAT)
- Support Federated Learning with 3rd parties
- Research and development of clinically impactful benchmarks (e.g., health equity, bias, dataset diversity)
- Co-develop and support benchmarking efforts
Comprehensive Open Federated Ecosystem (COFE)
- COFE is a collection of open tools that enables research and development of AI models within the clinical setting in efficient way. It consists of GaNDLF, MedPerf and OpenFL. With COFE researchers are able to design and run AI experiments in federated setting unlocking the power of diverse real-world clinical data.
Meeting Schedule
Wednesday October 29, 2025 Monthly – 10:05 – 11:00 Pacific Time
Medical Working Group Projects
How to Join and Access Medical AI Resources
To sign up for the group mailing list, receive the meeting invite, and access shared documents and meeting minutes:
- Fill out our subscription form and indicate that you’d like to join the Medical Working Group.
- Associate a Google account with your organizational email address.
- Once your request to join the Medical Working Group is approved, you’ll be able to access the Medical folder in the Public Google Drive.
To engage in working group discussions, join the group’s channels on the MLCommons Discord server.
To access the GitHub repositories (public):
- If you want to contribute code, please submit your GitHub ID to our subscription form.
- Visit the GitHub repositories:
Standards
MedPerf
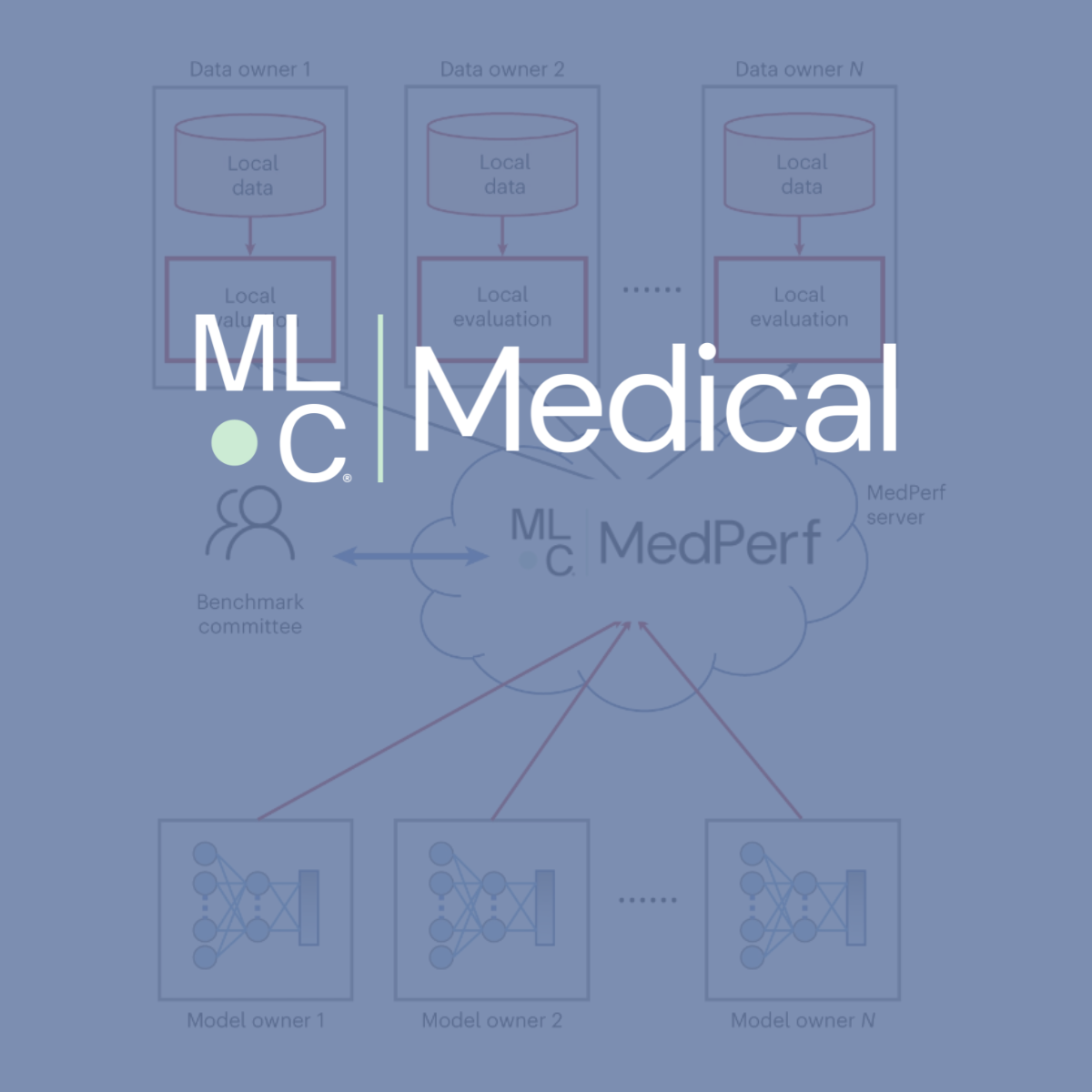
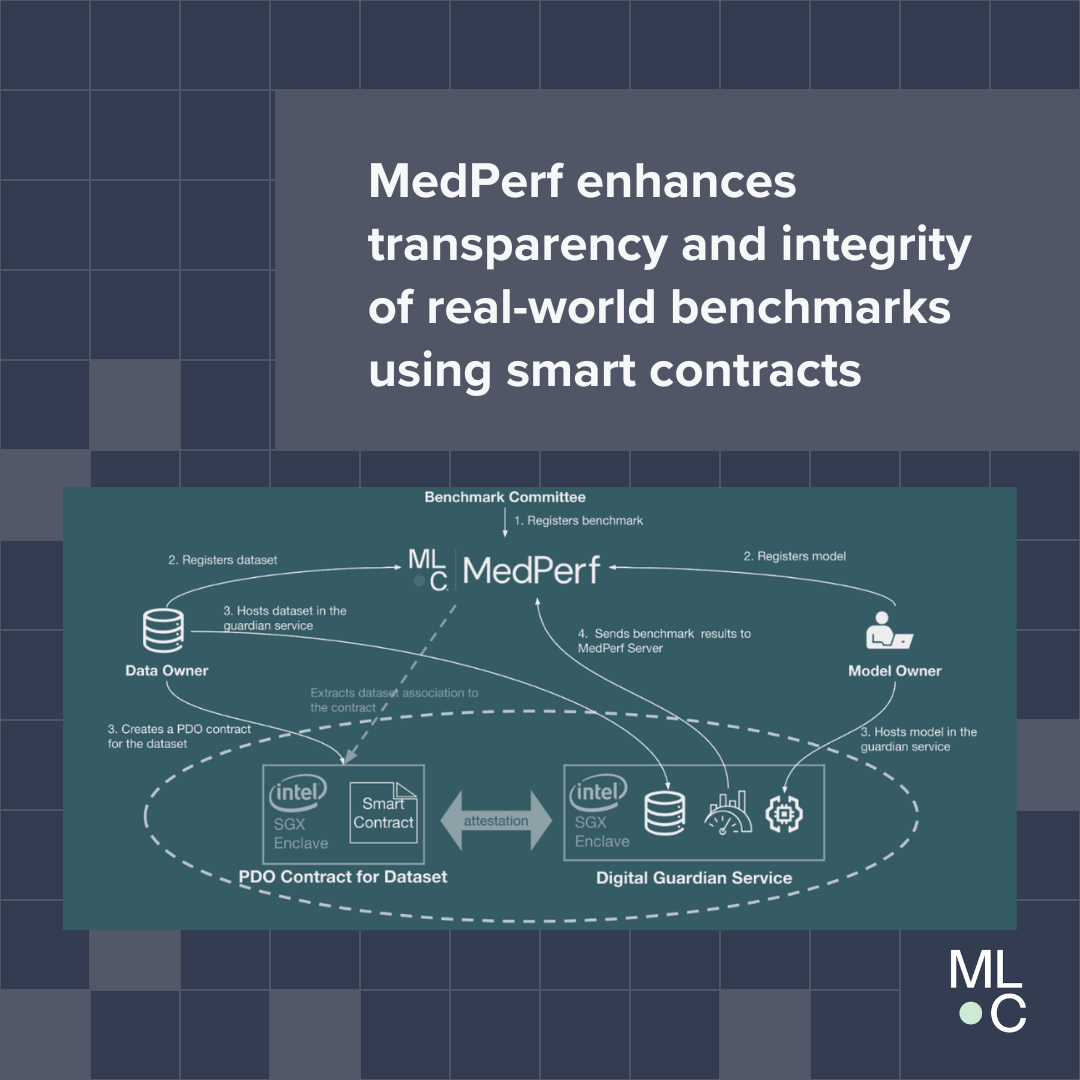
MedPerf is an open benchmarking platform that aims at evaluating AI on real world medical data. MedPerf follows the principle of federated evaluation in which medical data never leaves the premises of data providers. Instead, AI algorithms are deployed within the data providers and evaluated against the benchmark. Results are then manually approved for sharing with the benchmark authority. This effort aims to establish global federated datasets and to develop scientific benchmarks reducing risks of medical AI such as bias, lack of generalizability, and potential misuse. We believe that this two fold strategy will enable clinically impactful AI and drive healthcare efficacy. More information can be found at the MedPerf website and GitHub repository.
GaNDLF
GaNDLF is an open framework for developing machine learning workflows focused on healthcare. It follows zero/low code design principle and allows researchers to train and infer robust AI models without having to write a single line of code. Our effort aims to democratize medical AI through easy to use interfaces, well-validated machine learning ideologies, robust software development strategies, and a modular architecture to ensure integration with multiple open-source efforts. By focusing on end-to-end clinical workflow definition and deployment, we believe GaNDLF will turbocharge the AI landscape in healthcare. More information can be found at the GaNDLF website, GitHub repository, and paper.
Benchmarking of Medical AI
Call for Participation
We cannot achieve our goals without the help of the broader technical and medical community. We call for the following:
- Healthcare stakeholders to form benchmark committees that define specifications and oversee analyses.
- Participation of patient advocacy groups in the definition and dissemination of benchmarks.
- AI researchers, to test the end-to-end platform and use it to create and validate their own models across multiple institutions around the globe.
- Data owners (e.g., healthcare organizations, clinicians) to register their data in the platform (while never sharing the data).
- Data model standardization efforts to enable collaboration between institutions, such as the OMOP Common Data Model, possibly leveraging the highly multimodal nature of biomedical data.
- Regulatory bodies to develop medical AI solution approval requirements that include technically robust and standardized guidelines.
To-date we have worked with professionals from multiple hospitals, companies, universities, and groups:
A*STAR, Singapore • Amazon, Seattle, WA, USA • Brigham and Women’s Hospital, Boston, MA, USA • Broad Institute of MIT and Harvard, Cambridge, MA, USA • Cisco, San Jose, CA, USA • cKnowledge, Paris, France • Dana-Farber Cancer Institute, Boston, MA, USA • Factored, Palo Alto, CA, USA • Fast.ai, San Francisco, CA, USA • Flower Labs, Hamburg, Germany • Fondazione Policlinico Universitario A. Gemelli IRCCS, Rome, Italy • German Cancer Research Center, Heidelberg, Germany • Google, Mountain View, CA, USA • Harvard Medical School, Boston, MA, USA • Harvard T.H. Chan School of Public Health, Boston, MA, USA • Harvard University, Cambridge, MA, USA • Hugging Face, New York, NY, USA • IBM Research, San Jose, CA, USA • IHU Strasbourg, Strasbourg, France • Intel, Santa Clara, CA, USA • John Snow Labs, Lewes, DE, USA • Landing.AI, Palo Alto, CA, USA • Lawrence Livermore National Laboratory, Livermore, CA, USA • Massachusetts Institute of Technology, Cambridge, MA, USA • Meta, Menlo Park, CA, USA • Microsoft, Redmond, WA, USA • MLCommons, San Francisco, CA, USA • NVIDIA, Santa Clara, CA, USA • Nutanix, San Jose, CA, USA • OctoML, Seattle, WA, USA • Perelman School of Medicine, Philadelphia, PA, USA • Red Hat, Raleigh, NC, USA • Rutgers University, New Brunswick, NJ, USA • Sage Bionetworks, Seattle, WA, USA • Stanford University School of Medicine, Stanford, CA, USA • Stanford University, Stanford, CA, USA • Supermicro, San Jose, CA, USA • Tata Medical Center, Kolkata, India • University of Cambridge, Cambridge, UK • University of Heidelberg, Heidelberg, Germany • University of Pennsylvania, Philadelphia, PA, USA • University of Queensland, Brisbane, Australia • University of Strasbourg, Strasbourg, France • University of Toronto, Toronto, Canada • University of Trento, Trento, Italy • University of York, York, UK • Vector Institute, Toronto, Canada • Weill Cornell Medicine, New York, NY, USA • Write Choice, Florianópolis, Brazil
Call for Participation
We cannot achieve our goals without the help of the broader technical and medical community. We call for the following:
- Healthcare stakeholders to form benchmark committees that define specifications and oversee analyses.
- Participation of patient advocacy groups in the definition and dissemination of benchmarks.
- AI researchers, to test the end-to-end platform and use it to create and validate their own models across multiple institutions around the globe.
- Data owners (e.g., healthcare organizations, clinicians) to register their data in the platform (while never sharing the data).
- Data model standardization efforts to enable collaboration between institutions, such as the OMOP Common Data Model, possibly leveraging the highly multimodal nature of biomedical data.
- Regulatory bodies to develop medical AI solution approval requirements that include technically robust and standardized guidelines.
Medical AI Working Group Chairs
To contact all Medical AI working group chairs email [email protected].
Alexandros Karargyris
Renato Umeton
Vice Chairs
Micah Sheller
Vice Chair for System Architecture
Spyridon Bakas
Vice Chair for Benchmarking & Clinical Translation
Sarthak Pati
Vice Chair for Algorithm Development and Benchmarking